- Kirk Roberts, School of Biomedical Informatics, University of Texas Health Science Center, USA
- Parminder Bhatia, Amazon Health AI, USA
- Irene Spasic, School of Computer Science & Informatics, co-Director of the Data Innovation Research Institute, Cardiff University, UK
- Tristan Naumann, Microsoft Research Healthcare NExT, USA
- Carlos Luis Parra Calderón, Head of Technological Innovation, Virgen del Rocío University Hospital, Institute of Biomedicine of Seville, Spain
- Ashish Tendulkar, Google Research
- Alfonso Valencia Herrera, Barcelona Supercomputing Center (BSC-CNS), Spain
- Hercules Dalianis, Department of Computer and Systems Sciences, Stockholm University, Sweden
- Kevin Bretonnel Cohen, Colorado School of Medicine, USA; LIMSI, CNRS, Université Paris-Saclay, France
- Karin Verspoor, School of Computing and Information Systems, Health and Biomedical Informatics Centre, University of Melbourne, Australia
- Aurélie Névéol, LIMSI-CNRS, Université Paris-Sud, France
- Goran Nenadic, Department of Computer Science, University of Manchester
- Antonio Martinez, Head Pathology, Director National EQAS GCP, Spanish Society of Pathology, SEAP-IAP
- Zhiyong Lu, Deputy Director for Literature Search, National Center for Biotechnology Information (NCBI)
- Mauro Oruezabal, Head of Medical Oncology Service, Hospital Universitario Rey Juan Carlos, Madrid, Spain
Task Organizers
- Martin Krallinger, Text Mining Unit, Barcelona Supercomputing Center, Spain
- Antonio Miranda, Text Mining Unit, Barcelona Supercomputing Center, Spain
- Eulàlia Farré, Text Mining Unit, Barcelona Supercomputing Center, Spain
- Jose Antonio Lopez-Martin, Medical Oncology, Hospital Universitario 12 de Octubre; Instituto de Investigación Hospital 12 de Octubre (i+12), Spain
Description of the Corpus
Annotation guidelines can be downloaded from Zenodo.
Cantemist train, development, test and background sets are already available at Zenodo
Corpus format
- Subtasks CANTEMIST-NER: Brat annotation format.

Figure 1. Example Brat annotation for Cantemist-Ner.
- CANTEMIST-NORM: Brat annotation format.
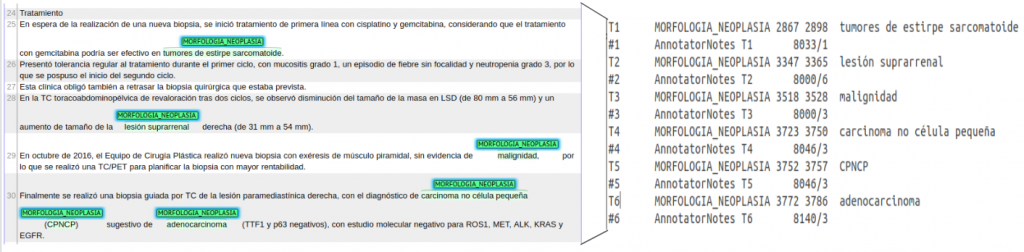
Figure 2. Example Brat annotation for Cantemist-Norm.
- Subtask CANTEMIST-CODING: CodiEsp format. We provide a single plain text file per clinical case and a tab-separated file with all the unique codes per clinical case (see Figure 3).
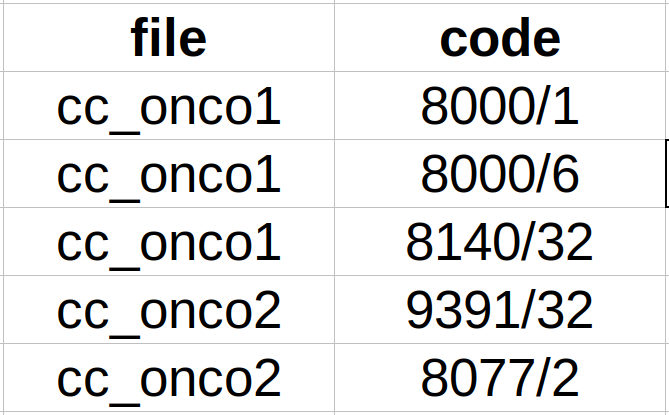
Figure 3. Example tab-separated file for Cantemist-Coding.
General information
For this task professional clinical coding experts have annotated a corpus of clinical cases in Spanish with eCIE-O-3.1 codes using the BRAT annotation tool following well-defined annotation guidelines adapted form the clinical coding recommendations published by the Spanish Ministry of Health, after several cycles of quality control and annotation consistency analysis before annotating the entire dataset. Figure 2 shows a screenshot of a sample manual annotation generated using the BRAT annotation tool.

Figure 2. Example BRAT annotation with labeled tumor morphology entity mention.
The CANTEMIST corpus consists of a collection of 3000 clinical cases that will be distributed in plain text in UTF8 encoding, where each clinical case would be stored as a single file. These clinical case reports were carefully selected to represent records reflecting as much as possible clinical narrative related to electronic clinical reports. Figure 3 illustrates an example text snippet corresponding to a short sample record.

Figure 3. Example plain text CANTEMIST corpus document
Additionally, we will also provide the annotation files comprising the character offsets of the tumor morphology entity mentions in TSV (tab-separated values) BRAT format together with their corresponding eCIE-O-3.1 code annotations.
The final corpus will be randomly split into three subsets: training, development and test. In the case of training and development sets, additionally, to the clinical cases, a TSV file will be released. It will contain one row per annotation. Each row will consist of the eCIE-O-3.1 code of the clinical case, a label indicating the category of the annotation, the annotation code and a reference to the text span that stimulated the annotation (the evidence).
In addition to the test set, a larger background set of clinical case documents will be released to make sure that participating teams will not be able to do manual corrections. In addition, the background set will become a silver standard of texts coded through automatic eCIE-O-3.1 code predictions returned by participating teams.
The goal of the CANTEMIST task is to develop automatic eCIE-O-3.1 clinical coding systems for Spanish medical texts. These systems should rely on the use of the CANTEMIST corpus, a high-quality Gold Standard synthetic clinical corpus of 3000 records based on a manual annotation process done by human clinical coding experts together with an inter-annotator agreement consistency analysis.
The CANTEMIST task can be approached as a named entity recognition and normalization task, but also as a multi-class text classification task. Participants are encouraged to either propose solutions in one of these directions or to combine both approaches. As well, novel approaches are welcomed.
Publications
All proceedings available at: http://ceur-ws.org/Vol-2664/
- Overview paper:
Named Entity Recognition, Concept Normalization and Clinical Coding: Overview of the Cantemist Track for Cancer Text Mining in Spanish, Corpus, Guidelines, Methods and Results. Antonio Miranda-Escalada, Eulàlia Farré-Maduell, Martin Krallinger. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 303-323 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_overview.pdf - Extracting Neoplasms Morphology Mentions in Spanish Clinical Cases through Word Embeddings. Pilar López-Úbeda, Manuel Carlos Díaz-Galiano, María Teresa Martín-Valdivia, Luis Alfonso Ureña-López. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 324-334 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper1.pdf - NLNDE at CANTEMIST: Neural Sequence Labeling and Parsing Approaches for Clinical Concept Extraction. Lukas Lange, Xiang Dai, Heike Adel, Jannik Strötgen. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 335-346 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper2.pdf - NCU-IISR: Pre-trained Language Model for CANTEMIST Named Entity Recognition. Jen-Chieh Han, Richard Tzong-Han Tsai. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 347-351 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper3.pdf - Recognai’s Working Notes for CANTEMIST-NER Track. David Carreto Fidalgo, Daniel Vila-Suero, Francisco Aranda Montes. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 352-357 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper4.pdf - End-to-End Neural Coder for Tumor Named Entity Recognition. Mohammed Jabreel. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 358-367 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper5.pdf - Using Embeddings and Bi-LSTM+CRF Model to Detect Tumor Morphology Entities in Spanish Clinical Cases. Sergio Santamaria Carrasco, Paloma Martínez. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 368-375 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper6.pdf - Tumor Entity Recognition and Coding for Spanish Electronic Health Records. Fadi Hassan, David Sánchez, Josep Domingo-Ferrer. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 376-384 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper7.pdf - Deep Neural Model with Contextualized-word Embeddings for Named Entity Recognition in Spanish Clinical Text. Renzo Rivera-Zavala, Paloma Martinez. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 385-395 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper8.pdf - Exploring Deep Learning for Named Entity Recognition of Tumor Morphology Mentions. Gema de Vargas Romero, Isabel Segura-Bedmar. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 396-411 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper9.pdf - Tumor Morphology Mentions Identification Using Deep Learning and Conditional Random Fields. Utpal Kumar Sikdar, Björn Gambäck, M Krishna Kumar. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 412-421 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper10.pdf - LasigeBioTM at CANTEMIST: Named Entity Recognition and Normalization of Tumour Morphology Entities and Clinical Coding of Spanish Health-related Documents. Pedro Ruas, Andre Neves, Vitor D.T. Andrade and Francisco M. Couto, Mario Ezra Aragón. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 422-437 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper11.pdf - A Parallel-Attention Model for Tumor Named Entity Recognition in Spanish. Tong Wang, Yuanyu Zhang, Yongbin Li. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 438-446 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper12.pdf - A Tumor Named Entity Recognition Model Based on Pre-trained Language Model and Attention Mechanism. Xin Taou, Renyuan Liu, Xiaobing Zhou. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 447-457 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper13.pdf - Identification of Cancer Entities in Clinical Text Combining Transformers with Dictionary Features. John D Osborne, Tobias O’Leary, James Del Monte, Kuleen Sasse. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 458-467 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper14.pdf - ICB-UMA at CANTEMIST 2020: Automatic ICD-O Coding in Spanish with BERT. Guillermo López-García, José Manuel Jerez, Nuria Ribelles, Emilio Alba, Francisco Javier Veredas. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 468-476 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper15.pdf - Automatic ICD Code Classification with Label Description Attention Mechanism. Kathryn Chapman, Günter Neumann. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 477-488 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper16.pdf - Vicomtech at CANTEMIST 2020. Aitor García-Pablos, Naiara Perez, Montse Cuadros. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 489-498 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper17.pdf - A Joint Model for Medical Named Entity Recognition and Normalization. Ying Xiong, Yuanhang Huang, Qingcai Chen, Xiaolong Wang, Yuan Nic, Buzhou Tang. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 499-504 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper18.pdf - Clinical NER using Spanish BERT Embeddings. Ramya Vunikili, Supriya H N, Vasile George Marica, Oladimeji Farri. Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings. 505-511 (2020).
URL: http://ceur-ws.org/Vol-2664/cantemist_paper19.pdf
The proceedings of the previous IberLEF2019 are online at http://ceur-ws.org/Vol-2421/
Working notes format
The working notes style is available via our proceedings volume template at
http://ceur-ws.org/Vol-XXX/ (we will use single-column format as in previous years).
Overleaf users can clone the style from
https://www.overleaf.com/read/gwhxnqcghhdt
Offline versions for LaTeX and DOCX are available from”
http://ceur-ws.org/Vol-XXX/CEURART.zip
Additionally we plan to prepare a journal special issue on the CANTEMIST task overview, corpus and results together with participating technical team systems descriptions in a Q1 journal.
Workshop
Cantemist (CANcer TExt Mining Shared Task) will be part of the IberLEF (Iberian Languages Evaluation Forum) 2020 evaluation campaign at the SEPLN 2020
36th Annual SEPLN Congress (September 23rd to 25th 2020, Málaga)
IberLEF aims to foster the research community to define new challenges and obtain cutting-edge results for the Natural Language Processing community, involving at least one of the Iberian languages: Spanish, Portugueses, Catalan, Basque or Galician. Accordingly, several shared-tasks challenges are proposed.
IberLEF 2020: https://sites.google.com/view/iberlef2020/
SEPLN 2020: http://sepln2020.sepln.org/index.php/en/iberlef-en/
Register for IberLEF and SEPLN here: http://sepln2020.sepln.org/index.php/registro/
All Cantemist talks will be available on YouTube: https://www.youtube.com/playlist?list=PL5uSCzf1azhC24g5dsp5eVMp8BZFWCraX.
| Local Time (CEST) - September 22nd, 2020 | Title | Presenter | Affiliation | More info |
|---|---|---|---|---|
| 6:40 pm | Named Entity Recognition, Concept Normalization and Clinical Coding: Overview of the Cantemist Track for Cancer Text Mining in Spanish, Corpus, Guidelines, Methods and Results | Antonio Miranda-Escalada | Barcelona Supercomputing Center, Spain | TBD |
| 7:00 pm | A Joint Model for Medical Named Entity Recognition and Normalization | Ying Xiong | Xili university town, China | TBD |
| 7:05 pm | Vicomtech at CANTEMIST 2020 | Naiara Pérez | Vicomtech, Spain | TBD |
| 7:10 pm | Extracting Neoplasms Morphology Mentions in Spanish Clinical Cases through Word Embeddings | Pilar López-Úbeda | University of Jaén, Spain | video |
| 7:15 pm | NLNDE at CANTEMIST: Neural Sequence Labeling and Parsing Approaches for Clinical Concept Extraction | Lukas Lange | Bosch Center for Artificial Intelligence, Germany | TBD |
| 7:20 pm | Tumor Entity Recognition and Coding for Spanish Electronic Health Records | Fadi Hassan | Universitat Rovira i Virgili, Spain | TBD |
| 7:25 pm | ICB-UMA at CANTEMIST 2020: Automatic ICD-O Coding in Spanish with BERT | Guillermo López-García | Universidad de Málaga, Spain | TBD |
| 7:30 pm | Conclusions and wrapup | Antonio Miranda-Escalada | Barcelona Supercomputing Center, Spain | - |
FAQ
Email Martin Krallinger to: encargo-pln-life@bsc.es
- Q: What is the goal of the shared task?
The goal is to predict the annotations (or codes) of the documents in the test and background sets. - Q: How do I register?
Here: https://temu.bsc.es/cantemist/?p=3956 - Q: How to submit the results?
We will provide further information in the following days.
Download the example ZIP file.
See Submission page for more info. - Q: Can I use additional training data to improve model performance?
Yes, participants may use any additional training data they have available, as long as they describe it in the working notes. We will ask to summarize such resources in your participant paper. - Q: The task consists of three sub-tasks. Do I need to complete all sub-tasks? In other words, If I only complete a sub-task or two sub-tasks, is it allowed?
Sub-tasks are independent and participants may participate in one, two or the three of them. - Q: How can I submit my results? Can I submit several prediction files for each sub-task?
You will have to create a ZIP file with your predictions file and submit it to EasyChair (further details will be soon released).
Yes, you can submit up to 5 prediction files, all in the same ZIP.
Download the example ZIP file.
See Submission page for more info. - Q: Should prediction files have headings?
No, prediction files should have no headings. - Q: Are all codes and mentions equally weighted?
Yes. However, systems will be evaluated including and excluding 8000/6 mentions. - Q: What version of the eCIE-O-3-1 is in use?
We are using the 2018 version. The table you can download from the official Spanish webpage is not complete. CIE-O allows combining the digits 6th and 7th according to the pathological study and the differentiation degree. That is, not all the combinations of the 6th and 7th characters are shown in the table.
There is a complete list of the valid codes on our webpage. Codes not present in this list will not be used for the evaluation. - Q. What is meant by the /H appended to various codes?
Some tumor mentions contain a relevant modifier not included in the terminology for this concept. Then, we append /H to the code.
For example, in the file cc_onco158, we have the codes 8000/1 and 8000/1/H.
8000/1 corresponds to a mention of neoplasm (“neoplasia”, in Spanish).
In the 8000/1/H case, the mention is (in Spanish) “neoplasia de estirpe epitelial”. The modifier “estirpe epitelial” is present in the ICD-O terminology for many tumors. However, it is not present to modify specifically the code 8000/1. Then, we consider it a relevant modifier and add the /H.
Schedule
| Event | Date | Link |
|---|---|---|
| Sample Set release | April, 28 | Sample set |
| Train Set Release and guidelines publication | June, 5 | Dataset and annotation guidelines |
| Development Set Release | June, 12 | Dataset |
| Test and Background Set Release | July, 3 | Dataset |
| End of evaluation period. Predictions submission deadline | August, 5, 23:59 CEST | Submission tutorial |
| Evaluation delivery and Test Set with Gold Standard annotations | August, 7 | Dataset |
| Working Notes deadline | August, 14, 23:59 CEST | Easychair |
| Working Notes Corrections deadline | August, 25 | |
| Camera-ready submission deadline | September, 1 | |
| IberLEF @ SEPLN 2020 | September, 22, from 16h to 20h | IberLEF |
Resources
Evaluation Script
- Official evaluation script: Available on GitHub (beta version).
This is the official evaluation script of the task.
Word embeddings
- Spanish Medical Word Embeddings. Word embeddings generated from Spanish medical corpora. Download them from Zenodo.
It can be used as a building block for clinical NLP systems used in Spanish texts.
Baseline
Dictionary lookup based on Levenshtein distance. It looks for train and development annotations in the test set.
Results (precision, recall, f1):
- NER: 0.181, 0.737, 0.291
- NORM: 0.18, 0.73, 0.288
- CODING (MAP): 0.584
Linguistic Resources
- CUTEXT. See it on GitHub.
Medical term extraction tool.
It can be used to extract relevant medical terms from clinical cases. - SPACCC POS Tagger. See it on Zenodo.
Part Of Speech Tagger for Spanish medical domain corpus.
It can be used as a component of your system. - NegEx-MES. See it on Zenodo.
A system for negation detection in Spanish clinical texts based on NegEx algorithm.
It can be used as a component of your system. - Negation corpus. See it on GitHub
A Corpus of Negation and Uncertainty in Spanish Clinical Texts (and instructions to train the system). - AbreMES-X. See it on Zenodo.
Software used to generate the Spanish Medical Abbreviation DataBase. - AbreMES-DB. See it on Zenodo.
Spanish Medical Abbreviation DataBase.
It can be used to fine-tune your system. - MeSpEn Glossaries. See it on Zenodo.
Repository of bilingual medical glossaries made by professional translators.
It can be used to fine-tune your system.
Terminological Resources
- List of valid codes. Download it from here.
List of valid ICD-O-3 codes used in the task evaluation.
Other Relevant Systems
Evaluation
Evaluation will be done by comparing the automatically generated results to the results generated by manual annotation of experts.
The primary evaluation metric for all three sub-tracks will consist of micro-averaged precision, recall and F1-scores:

The used evaluation scripts together with a Readme file with instructions will be available on GitHub to enable systematic fine-tuning and improvement of results on the provided training/development data using by participating teams.
For the CANTEMIST-CODING sub-track we also apply a standard ranking metric: Mean Average Precision (MAP) for evaluation purposes.
MAP (Mean Average Precision) is an established metric used for ranking problems.
All metrics will be computed including and excluding mentions with 8000/6 code.
Examples
Examples are taken from the evaluation library toy data, available on GitHub
Example 1: Evaluate the toy data output for CANTEMIST-NER
$> cd src $> python main.py -g ../gs-data/ -p ../toy-data/ -s ner ----------------------------------------------------- Clinical case name Precision ----------------------------------------------------- cc_onco1.ann 0.5 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average precision = 0.846 ----------------------------------------------------- Clinical case name Recall ----------------------------------------------------- cc_onco1.ann 0.667 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average recall = 0.917 ----------------------------------------------------- Clinical case name F-score ----------------------------------------------------- cc_onco1.ann 0.571 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average F-score = 0.88
Example 2: Evaluate the toy data output for CANTEMIST-NORM
$> cd src $> python main.py -g ../gs-data/ -p ../toy-data/ -s norm ----------------------------------------------------- Clinical case name Precision ----------------------------------------------------- cc_onco1.ann 0.25 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average precision = 0.769 ----------------------------------------------------- Clinical case name Recall ----------------------------------------------------- cc_onco1.ann 0.333 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average recall = 0.833 ----------------------------------------------------- Clinical case name F-score ----------------------------------------------------- cc_onco1.ann 0.286 ----------------------------------------------------- cc_onco3.ann 1.0 ----------------------------------------------------- Micro-average F-score = 0.8
Example 3: Evaluate the toy data output for CANTEMIST-CODING
$> cd src $> python main.py -g ../gs-data/gs-coding.tsv -p ../toy-data/pred-coding.tsv -c ../valid-codes.tsv -s coding MAP estimate: 0.75