Submission instructions
5 submissions per sub-track will be allowed.
You must submit ONE SINGLE ZIP file with the following structure:
- One subdirectory per subtask in which you are participating.
- In addition, in the parent directory, you must add a README.txt file with your contact details and a really short explanation of your system.
- If you have more than one system, you can include their predictions and we will evaluate them (up to 5 prediction runs).
- Cantemist-NER and Cantemist-NORM:
- You must include the Brat annotation files (.ANN) with your predictions.
- One annotation file per document.
- If you have more than one system, create sub-directories inside the cantemist-ner and cantemist-norm directories. One subdirectory per system.
- If you have more than one system, name the subdirectories with numbers and a recognizable name. For example, 1-systemDL and 2-systemlookup
- Cantemist-CODING:
- You must include the tab-separated file with your predictions.
- One single file with all the predictions.
- With a .tsv file extension.
- If you have more than one system, include one tab-separated file for each system.
- If you have more than one system, name the tab-separated files with numbers and a more or less recognizable name. For example, 1-systemDL.tsv and 2-systemlookup.tsv
Download here a submission ZIP example.
Submission method
Submissions will be made via SFTP.
Download here the submission tutorial.
Submission format
- CANTEMIST-NER
Brat Format: one ANN file per document. ANN files have the following format:
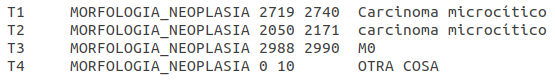
- CANTEMIST-NORM
Brat Format: one ANN file per document. ANN files have the following format (with the codes added as Brat comments):

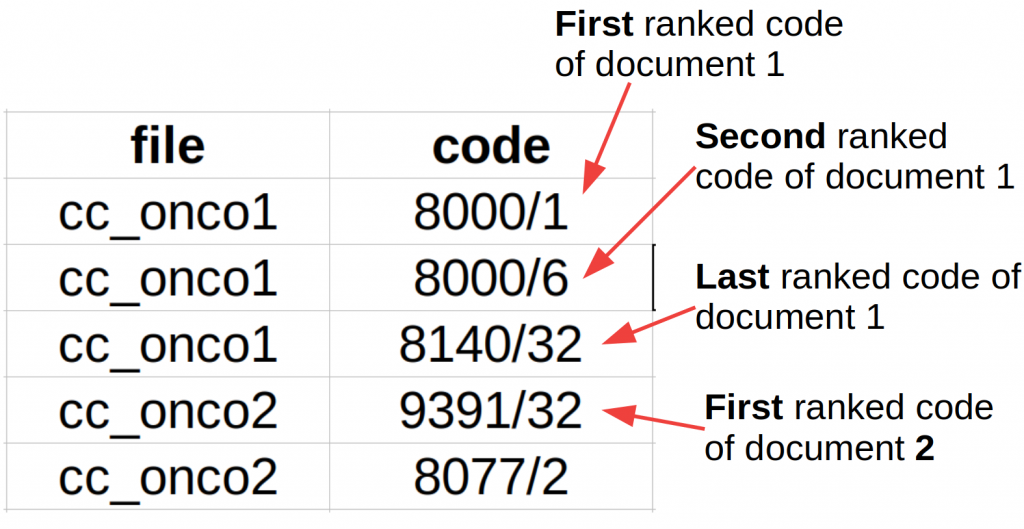
- CANTEMIST-CODING
A tab-separated with two columns: clinical case and code. Codes must be ordered by rank/confidence, with more relevant codes first. For example: