ClinSpEn
This website contains the data for the ClinSpEn shared task, focused on clinical EN-ES machine translation and part of the biomedical task of WMT 2022.
The Gold Standard test sets for every dataset have been released. They are available on Zenodo.
The BioWMT 2022 overview paper is now available! You can find it here. If you use any of the ClinSpEn data, please remember to cite it.
Motivation
Machine translation applied to the clinical domain is a specially challenging task due to the complexity of medical language and the heavy use of health-related technical terms and medical expressions. Therefore there is a large community of specialized medical translators, able to deal with medical narratives, terminologies or the use of ambiguous abbreviations and acronyms.
Taking into account the relevance, impact and diversity of health-related content, as well as the rapidly growing number of publications, EHRs, clinical trials, informed consent documents and medical terminologies there is a pressing need to be able to generate more robust medical machine translation resources together with independent quality evaluation scenarios.
Recent advances in machine translation technologies together with the use of other NLP components are showing promising results, thus domain adaptation of MT approaches can have a significant impact in unlocking key information from medical content.
Therefore, the ClinSpEn data represents three different types of data very relevant to the biomedical domain: clinical cases, clinical terminology and ontology concepts.
Sub-tracks
All in all, ClinSpEn is comprised of three different sub-tracks:
- ClinSpEn-CC (clinical cases): EN>ES translation of clinical cases using a collection of 202 parallel COVID-19 clinical case reports.
- ClinSpEn-CT (clinical terms): ES>EN translation of clinical terminology using a collection of over 19 000 parallel terms obtained from biomedical literature and electronic health records.
- ClinSpEn-OC (ontology concepts): EN>ES translation of a collection of over 2 000 parallel concepts obtained from different biomedical ontologies.
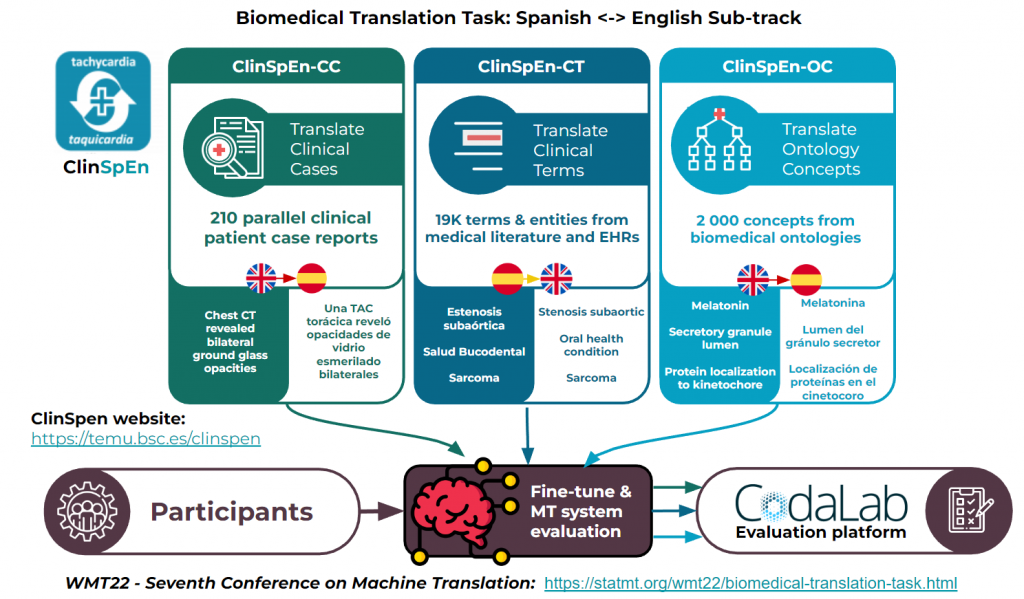
All documents and terms in the ClinSpEn collection have been manually translated and revised by professional medical translators in order to ensure the quality and validity of the data.
ClinSpEn is organized by the Barcelona Supercomputing Center’s NLP for Biomedical Information Analysis group (formerly Text Mining Unit).
Related resources
At the NLP for Biomedical Information Analysis group, one of our missions is the open publication of datasets to train and benchmark biomedical information extraction, normalization and indexing systems. For that reason, we have released multiple datasets as part of shared tasks over the years. If you are interested in ClinSpEn, you might want to take a look at some of our resources and competitions about:
- Clinical content extraction: MedProcNER (clinical procedures — new this year!), DisTEMIST (diseases), CANTEMIST (tumour morphology), CodiEsp (coding to ICD), PharmaCoNER (chemicals and proteins)
- Sociodemographic content extraction: MEDDOPLACE (locations, clinical departments and related info — new this year!), MEDDOCAN (sensitive data), MEDDOPROF (occupations)
- Information extraction in social media: SocialDisNER (diseases), ProfNER (occupations)
- Linguistic aspects: BARR1 and BARR2 (abbreviation resolution)