- Olivier Bodenreider, US National Library of Medicine, USA
- Kevin Cohen, University of Colorado School of Medicine, USA
- Robert Leaman, US National Library of Medicine, USA
- Diego Molla, Macquarie University, Australia
- Zhiyong Lu, US National Library of Medicine, USA
- Azadeh Nikfarjam, Apple, USA
- Thierry Poibeau, French National Center for Scientific Research, France
- Kirk Roberts, University of Texas Health Science Center at Houston, USA
- Yutaka Sasaki, Toyota Technological Institute, Japan
- H. Andrew Schwartz, Stony Brook University, USA
- Nicolas Turenne, French National Institute for Agricultural Research, France
- Karin Verspoor, University of Melbourne, Australia
- Pierre Zweigenbaum, French National Center for Scientific Research, France
Task Organizers
Social Media Mining for Health Applications (#SMM4H) Shared Task 2021 is organized by:
- Graciela Gonzalez-Hernandez, University of Pennsylvania, USA
- Davy Weissenbacher, University of Pennsylvania, USA
- Ari Z. Klein, University of Pennsylvania, USA
- Karen O’Connor, University of Pennsylvania, USA
- Abeed Sarker, Emory University, USA
- Elena Tutubalina, Kazan Federal University, Russia
- Martin Krallinger, Barcelona Supercomputing Center, Spain
- Anne-Lyse Minard, Université d’Orléans, France
ProfNER-ST is organized by:
- Antonio Miranda-Escalada, Barcelona Supercomputing Center, Spain
- Eulàlia Farré-Maduell, Barcelona Supercomputing Center, Spain
- Salvador Lima, Barcelona Supercomputing Center, Spain
- Vicent Briva-Iglesias, D-REAL, Dublin CityUniversity
- Martin Krallinger, Barcelona Supercomputing Center, Spain
Description of the Corpus
Training and validation (annotated), test and background (unannotated) datsets
The SMM4H-Spanish corpus is a collection of 10,000 health-related tweets in Spanish annotated with professions, employment statuses and other work-related activities. The aim of the corpus is to extract professions from social media to enable characterizing health-related issues, in particular in the context of COVID-19 epidemiology as well as mental health conditions.
The data of the corpus was obtained from a Twitter crawl that used keywords like “Covid-19”, “epidemia” (epidemic) or “confinamiento” (lockdown), as well as hashtags such as “#yomequedoencasa” (#istayathome), to retrieve relevant tweets. This crawl was further filtered to obtain only the tweets that were written both from Spain and in Spanish.
The corpus was annotated by linguist experts in an iterative process that included the creation of annotation guidelines specifically for this task. These guidelines are described and available for download here.
We have performed a consistency analysis of the corpus. 10% of the documents have been annotated by an internal annotator as well as by the linguist experts. The preliminary Inter-Annotator Agreement (pairwise agreement) is 0.919.
The annotation process was performed using the web-based tool brat. Below is an example of how the annotated tweets look like:

All in all, 10,000 tweets were annotated. They were split into 60% training (6,000), 20% development (2,000) and 20% test (2,000). The different splits can be downloaded here.
FORMAT
Track A – Tweet binary classification. Annotations are stored in a tab-separated file with 2 columns:
tweet_id label
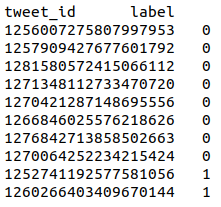
Track B – Tweet binary classification. Annotations are stored in a tab-separated file with 5 columns:
tweet_id begin end type extraction
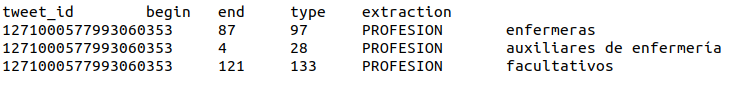
In addition, the corpus of Track B is provided in Brat format and in the IOB tagging scheme. See it on Zenodo.
Datasets
Training and validation (annotated), test and background (unannotated) datsets
Train set
The train set contains around 6,000 annotated tweets. Find it in Zenodo
Validation set
The validation set contains around 2,000 annotated tweets. Find it in Zenodo
Test and Background sets
The test set contains around 2,000 tweets. The background set contains 25K tweets. Find it in Zenodo.
You must submit predictions for the test and background sets. But you will only be evaluated for the test set predictions.
Test set with Gold Standard annotations
The Gold Standard annotations of the test set will be released after the submission deadline
Publications
ProfNER’s overview:
Miranda-Escalada, A., Farré-Maduell, E., Lima-López, S., Gascó, L., Briva-Iglesias, V., Agüero-Torales, M., & Krallinger, M. (2021, June). The ProfNER shared task on automatic recognition of occupation mentions in social media: systems, evaluation, guidelines, embeddings and corpora. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 13-20).
URL: https://aclanthology.org/2021.smm4h-1.3/
SMM4H 2021 overview paper:
Magge, A., Klein, A., Miranda-Escalada, A., Al-Garadi, M. A., Alimova, I., Miftahutdinov, Z., … & Gonzalez, G. (2021, June). Overview of the sixth social media mining for health applications (# smm4h) shared tasks at naacl 2021. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 21-32).
URL: https://aclanthology.org/2021.smm4h-1.4/
Participants’ papers:
- De Leon, F. A. L., Madabushi, H. T., & Lee, M. (2021, June). UoB at ProfNER 2021: Data Augmentation for Classification Using Machine Translation. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 115-117).
- Fidalgo, D. C., Vila-Suero, D., Montes, F. A., & Cepeda, I. T. (2021, June). System description for ProfNER-SMMH: Optimized finetuning of a pretrained transformer and word vectors. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 69-73).
- Ruas, P., Andrade, V., & Couto, F. M. (2021, June). Lasige-biotm at profner: Bilstm-crf and contextual spanish embeddings for named entity recognition and tweet binary classification. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 108-111).
- Yaseen, U., & Langer, S. (2021). Neural text classification and stacked heterogeneous embeddings for named entity recognition in smm4h 2021. arXiv preprint arXiv:2106.05823.
- Păiș, V., & Mitrofan, M. (2021, June). Assessing multiple word embeddings for named entity recognition of professions and occupations in health-related social media. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 128-130).
- Murgado, A. M., Portillo, A. P., Úbeda, P. L., Martín-Valdivia, M. T., & Lopez, L. A. U. (2021, June). Identifying professions & occupations in health-related social media using natural language processing. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 141-145).
- Carrasco, S. S., & Rosillo, R. C. (2021, June). Word embeddings, cosine similarity and deep learning for identification of professions & occupations in health-related social media. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 74-76).
- Pachón, V., Vázquez, J. M., & Olmedo, J. L. D. (2021, June). Identification of profession & occupation in health-related social media using tweets in spanish. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 105-107).
- Zhou, T., Li, Z., Gan, Z., Zhang, B., Chen, Y., Niu, K., … & Liu, S. (2021, June). Classification, extraction, and normalization: Casia_unisound team at the social media mining for health 2021 shared tasks. In Proceedings of the Sixth Social Media Mining for Health (# SMM4H) Workshop and Shared Task (pp. 77-82).
Workshop
Participating teams are required to submit a paper describing the system(s) they ran on the test data. Sample description systems can be found in pages 89-136 of the #SMM4H 2019 proceedings. Accepted system descriptions will be included in the #SMM4H 2021 proceedings.

We encourage, but do not require, at least one author of each accepted system description to register for the #SMM4H 2021 Workshop, co-located at “NAACL“, and present their system as a poster. Select participants, as determined by the program committee, will be invited to extend their system description to up to four pages, plus unlimited references, and present their system orally.
Contact & FAQ
Email Martin Krallinger to encargo-pln-life@bsc.es or Antonio Miranda to antonio.miranda@bsc.es
- Q: What is the goal of the shared task?
The goal is to predict the category (Track 1) or the named entities (Track 2) of the tweets in the test and background sets. - Q: How do I register?
Here: https://forms.gle/1qs3rdNLDxAph88n6 - Q: How do I submit the results?
In CodaLab: https://competitions.codalab.org/competitions/28766 - Q: Can I use additional training data to improve model performance?
Yes, participants may use any additional training data they have available, as long as they describe it in the system description. - Q: ProfNER has two tracks. Do I need to complete all tracks?
Sub-tracks are independent and participants may participate in one or two of them. - Q: Is there a Google Group for the ProfNER task?
Yes: https://groups.google.com/g/smm4h21-task-7
Schedule
TBD
Additional Resources
Evaluation Script
- Official evaluation script: TBD
Linguistic Resources
- CUTEXT. See it on GitHub.
Medical term extraction tool.
It can be used to extract relevant medical terms from tweets. - SPACCC POS Tagger. See it on GitHub.
Part Of Speech Tagger for Spanish medical domain corpus.
It can be used as a component of your system. - NegEx-MES. See it on Zenodo and on GitHub.
A system for negation detection in Spanish clinical texts based on NegEx algorithm.
It can be used as a component of your system. - AbreMES-X. See it on Zenodo.
Software used to generate the Spanish Medical Abbreviation DataBase. - AbreMES-DB. See it on Zenodo.
Spanish Medical Abbreviation DataBase.
It can be used to fine-tune your system. - MeSpEn Glossaries. See it on Zenodo.
Repository of bilingual medical glossaries made by professional translators.
It can be used to fine-tune your system. - Occupations gazetteer. See it on Zenodo.
A gazetter of occupations extracted from a set of terminologies (DeCS, ESCO, SnomedCT and WordNet) and Stanford CoreNLP.
Word embeddings
- FastText Spanish medical embeddings. See them on Zenodo.
Word and subword embeddings trained for medical Spanish domain.
It can be used as a component of your system. - FastText Spanish Twitter embeddings. See them on Zenodo.
Word and subword embeddings trained with Spanish Twitter data related to COVID-19.
It can be used as a component of your system.
Baseline
More resources TBD
Evaluation
The evaluation will be done at CodaLab
Precision (P) = true positives/(true positives + false positives)
Recall (R) = true positives/(true positives + false negatives)
F-score (F1) = 2*((P*R)/(P+R))
Track A – Tweet binary classification
Submissions will be ranked by Precision, Recall and F1-score for the positive class (F-score is the primary metric).
Prediction format: tab-separated file with headers:
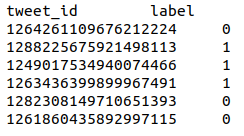
Track B – NER offset detection and classification
Submissions will be ranked by Precision, Recall and F1-score for each PROFESION [profession] or SITUACION_LABORAL [working status] mention extracted, where the spans overlap entirely (F-score is the primary metric).
A correct prediction must have the same beginning and ending offsets as the Gold Standard annotation, as well as the same label (PROFESION or SITUACION_LABORAL)
Prediction format: tab-separated file with headers. Same as the tab-separated format of the Gold Standard.

CodaLab
Predictions for each subtask should be contained in a single .tsv (tab-separated values) file. This file (and only this file) should be compressed into a .zip file. Please upload this zip file as submission. For the evaluation phase which will start on the 1st of March, you are allowed to add the validation set to the training set for training purposes.
Codalab: https://competitions.codalab.org/competitions/28766
- Register and wait for approval
- To make submissions : Participate -> Submit/View Results -> Click on Task -> Click Submit -> Select File
Refresh your submission. It goes from Submitted -> Running -> Finished. Scores should be available in the files. You can choose to submit your best scores to the Leaderboard. - To view results : Results -> Click on Task -> View results in table
You will be allowed to make unlimited submissions during the validation stage. During the evaluation stage only 2 submissions will be allowed.