Official Annotation Guidelines used to annotate the MEDDOCAN data sets can be downloaded from here.
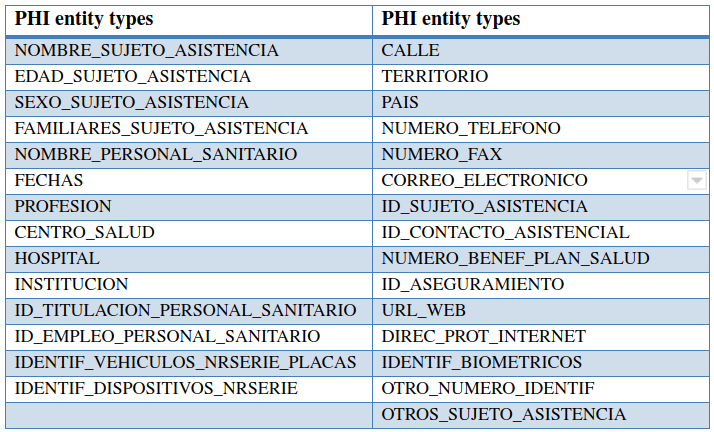
The MEDDOCAN annotation scheme defines a total of 29 granular entity types grouped into more general parent classes. Figure 1 summarized the list of sensitive entity types defined for the MEDDOCAN track.
The annotation process of the MEDDOCAN corpus was inspired initially by previous annotation schemes and corpora used for the i2b2 de-identification tracks, revising the guidelines used for these tracks, translating certain characteristics into Spanish and adapting them to the specificities and needs of our document collection and legislative framework. This adaptation was carried out in collaboration with practicing physicians, a team of annotators and the University Hospital 12 de Octubre. The adaptation, translation, and refinement of the guidelines was carried out on several random sample sets of the MEDDOCAN corpus and connected to an iterative process of annotation consistency analysis through inter-annotator agreement (IAA) calculation until a high annotation quality on terms of IAA was reached. Three cycles of refinement and IAA analysis were needed in order to reach the quality criteria required for this track, being in line with similar scores obtained for instance for i2b2. A link to the final version of the used 28 pages annotation guidelines can be downloaded from here.
This iterative refinement process required direct interaction between the expert annotators in order to resolve discrepancies, using a side-by-side visualization of clinical case document annotations, by comparing discrepancies and going over each of them in order to solve doubts, add/refine rules and add/edit clarifying examples to the guidelines. The final, inter-annotator agreement measure obtained for this corpus was calculated on a set of 50 records that were double annotated (blinded) by two different expert annotators, reaching a pairwise agreement of 98% on the exact entity mention comparison level together with the corresponding mention type labels.
The manual annotation of the entire corpus was carried out in a three-step approach. First, an initial annotation process was carried out on an adapted version of the AnnotateIt tool. The resulting annotations were then exported, trailing whitespaces were removed and double annotations of the same string were send as an alert to the human annotators for revision/correction. Then, the annotations were uploaded into the BRAT annotation tool, which was slightly less efficient for mention labeling in terms of manual annotation speed. The human annotators performed afterwards a final revision of the annotations in BRAT, in order to correct mistakes and to add potentially missing annotation mentions. Finally the senior annotator did a last round of annotation revision of the entire corpus. Figure 2 shows an example of an annotation in the BRAT format.