Submission
This page includes two parts:
Submission Instructions
To submit your results, (1) register at BioASQ website and (2), follow the following instructions:
A. Up to 5 submissions per sub-track will be allowed.
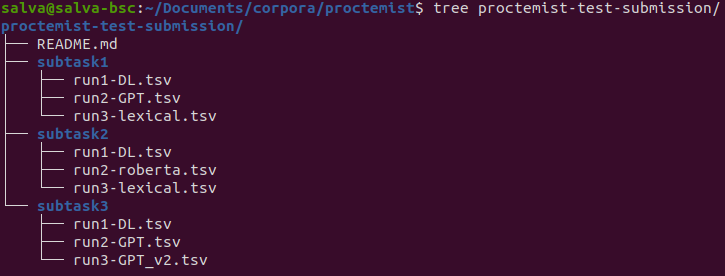
B. You must submit ONE SINGLE ZIP file with the following structure:
- One subdirectory per subtask in which you are participating.
- In addition, in the parent directory, you must add a README file with your contact details and a really short explanation of your system.
- If you have more than one system, you can include their predictions and we will evaluate them (up to 5 prediction runs).
- For each prediction run:
- You must include a single tab-separated file (.TSV) with your predictions.
- Your .TSV file must include headers (see Submission Instructions below to know which ones to add depending on the task)
- If you have more than one system, include one tab-separated file for each system.
- If you have more than one system, name the tab-separated files with numbers and a more or less recognizable name. For example, 1-systemDL.tsv and 2-systemlookup.tsv
- This is an example of what your .ZIP file should look like:
C. Once your .ZIP file is ready, access the BioASQ’s Participants Area, click on Submitting > Task MedProcNER, where you will be able to upload your predictions. Keep in mind that only one .ZIP file is allowed. Uploading a new one will replace the previous one.
Submission Instructions
All submissions must be done using .TSV files. These are the columns you must provide for each sub-track:
Sub-track 1 (MedProcNER-Entities)
- filename: Name of the file from which the procedure mention has been extracted.
- label: In this case it will always be PROCEDIMIENTO
- start_span: Character number where the detected mention starts.
- end_span: Character number where the detected mention ends.
- text: Mention extracted from text
Sub-track 2 (MedProcNER-Normalization)
- filename: Name of the file from which the procedure mention has been extracted.
- label: In this case it will always be PROCEDIMIENTO.
- start_span: Character number where the detected mention starts.
- end_span: Character number where the detected mention ends.
- text: Mention extracted from text.
- code: List of Snomed-CT concept codes linked to the mention. If there is more than one code associated with a mention, they will be concatenated by the symbol “+”.
Sub-track 3 (MedProcNER-Indexing)
- filename: Name of each file.
- code: List of Snomed-CT concept codes proposed to index the document. Different codes must be concatenated by the symbol “+”.