Download the corpus from zenodo
This page contains the following information:
1. General information
The LivingNER Gold Standard consists of a collection of 2000 clinical case reports that will be distributed in plain text in UTF8 encoding, where each clinical case would be stored as a single file. Additionally, we will also provide the annotation files comprising the character offsets of the entity mentions in TSV (tab-separated values) files together with their corresponding NCBI Taxonomy code annotations.

The clinical case reports come from 20 medical disciplines (enfermedades infecciosas (incluidos casos de Covid-19), cardiología, neurología, oncología, otorrinolaringología, odontología, pediatría, endocrinología, atención primaria, alergología, radiología, psiquiatría, oftalmología, psiquiatría, urología, medicina interna, emergencias y medicina de cuidados intensivos, radiología, medicina tropical y dermatología) with species [SPECIES] and [HUMAN] entities manually annotated.
The corpus’ content is quite varied, as it includes annotations for animals, plants, and microorganisms (including bacteria, fungi, viruses, and parasites). Both scientific names, as well as common names, were considered.
All of these mentions have been manually mapped to the NCBI taxonomy. Please, beware that:
- Composite mentions. If several NCBI taxonomy codes were required to map a single annotated mention, the codes are concatenated with a “|” symbol. For instance, “microorganism” is mapped to “2|2759|10239”.
- Terminology codes that are more general than the annotated mention. If the NCBI taxonomy concept was more general than the annotated mention, the modifier “H” is added to the NCBI taxonomy code. For instance, “baciloscopia” is mapped to “2|H”.
The final corpus will be randomly split into three subsets: training, development and test. In the case of training and development sets, additionally, to the clinical cases, a TSV file will be released. It will contain one row per annotation.
In addition to the test set, a larger background set of clinical case documents will be released to make sure that participating teams will not be able to do manual corrections.
The goal of the LivingNER task is to develop automatic systems for Spanish medical texts. These systems should rely on the use of the LivingNER corpus, a high-quality Gold Standard synthetic clinical corpus of 2000 records based on a manual annotation process done by human experts together with an inter-annotator agreement consistency analysis.
2. Corpus format
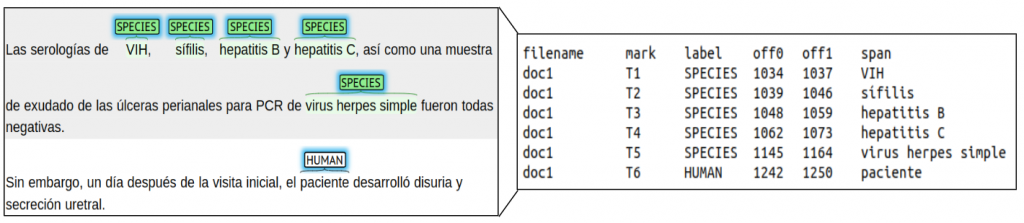
For subtask 1 (LivingNER – Species NER), annotations are distributed in a tab-separated file (TSV) file with the following columns:
- filename: document name
- mark: identifier mention mark
- label: mention type (SPECIES or HUMAN)
- off0: starting position of the mention in the document
- off1: ending position of the mention in the document
- span: textual span
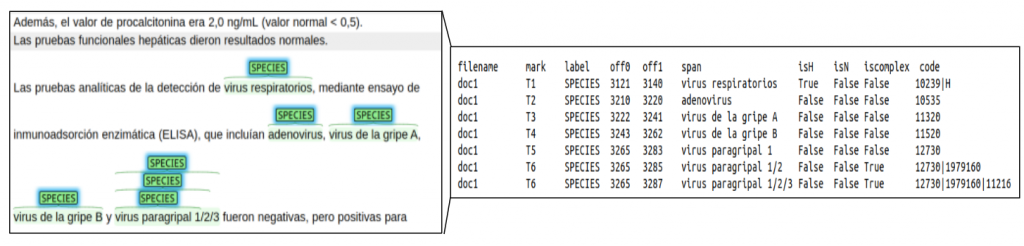
For subtask 2 (LivingNER – Species Norm), annotations are distributed in a TSV file with the same columns as the previous one, plus:
- isH: whether the span is narrower than the NCBITax assigned code
- isN: whether the mention corresponds to a nosocomial infection
- iscomplex: whether the span has assigned a combination of NCBITax codes
- NCBITax: mention code in the NCBI Taxonomy
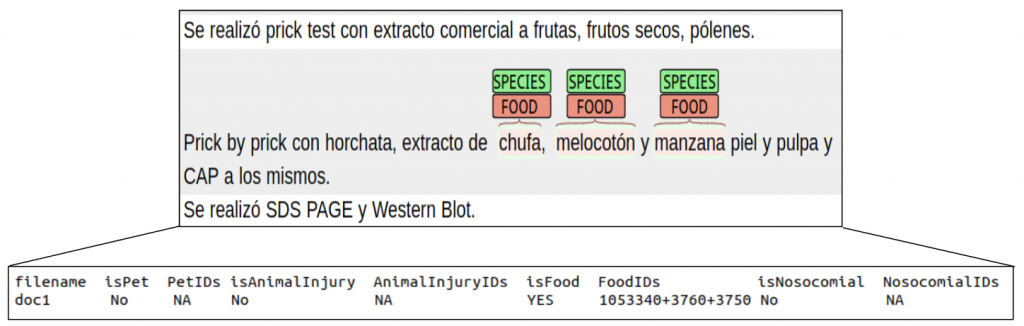
For subtask 3 (LivingNER – Clinical IMPACT), annotations are distributed in a (TSV) with the following columns:
- filename
- isPet (Yes/No)
- PetIDs (NCBITaxonomy codes of pet & farm animals present in document)
- isAnimalInjury (Yes/No)
- AnimalInjuryIDs (NCBITaxonomy codes of animals causing injuries present in document)
- IsFood (Yes/No)
- FoodIDs (NCBITaxonomy codes of food mentions present in document)
- isNosocomial (Yes/No)
- NosocomialIDs (NCBITaxonomy codes of nosocomial species mentions present in document)
All text files are distributed as plain UTF-8 text files, where each clinical case would be stored as a single file.