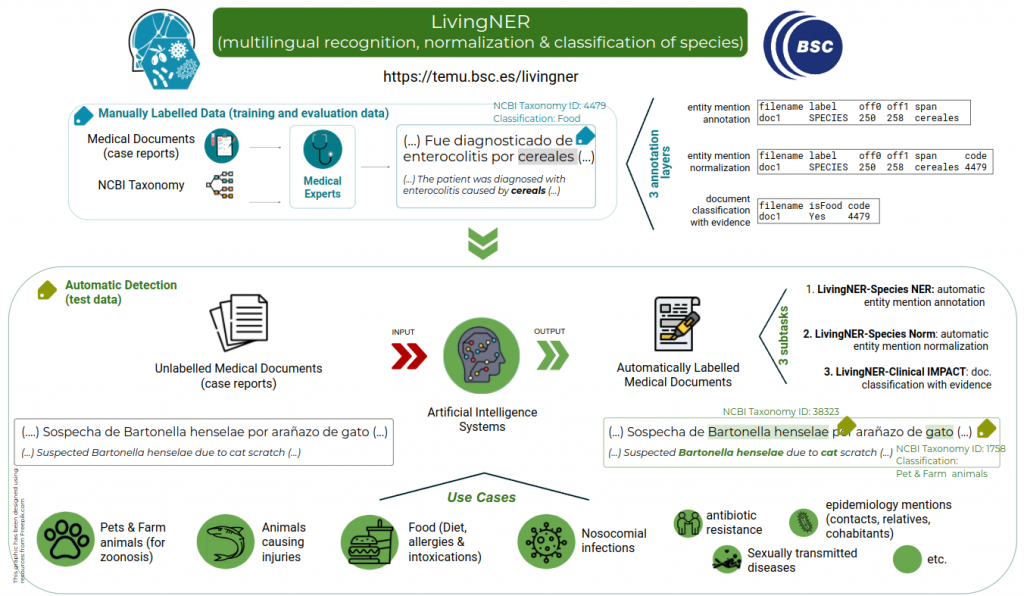
LivingNER: Named entity recognition, normalization & classification of species, pathogens and food
The LivingNER Track is promoted by Plan de Impulso de las Tecnologías del Lenguaje (Plan TL)
Generated resources
- Data: Gold Standard, Multilingual Corpus, Silver Standard & Annotation Guidelines
- Task Overview paper, Conference Proceedings
- YouTube presentation (TBA)
- Participant codes
* NEW February 28th 2023: The annotated Test Set has been publicly released
Please, cite us:
A. Miranda-Escalada, E. Farré-Maduell, S. Lima-López, D. Estrada, L. Gascó, M. Krallinger, Mention detection, normalization & classification of species, pathogens, humans and food in clinical documents: Overview of livingner shared task and resources, Procesamiento del Lenguaje Natural (2022)
@article{amiranda2022nlp,
title={Mention detection, normalization \& classification of species, pathogens, humans and food in clinical documents: Overview of LivingNER shared task and resources},
author={Miranda-Escalada, Antonio and Farr{\'e}-Maduell, Eul{`a}lia and Lima-L{\'o}pez, Salvador and Estrada, Darryl and Gasc{\'o}, Luis and Krallinger, Martin},
journal = {Procesamiento del Lenguaje Natural},
year={2022}
}
This page contains the following information:
- LivingNER initiative: Background and Motivation
- Task Summary
- LivingNER Gold Standard corpus
- Scientific Publications
IberLEF 2022 Workshop @ SEPLN 2022 (A Coruña)
1. Background and Motivation
Organisms/species have scarcely featured in NLP studies to date, in particular for non-English content. High-quality guidelines and corpora providing granular text-bound semantic annotations are needed to leverage mentions of living creatures in biomedical texts. The annotation of species or living organisms is critical to scientific disciplines like medicine, biology, ecology/biodiversity, nutrition and agriculture.
Hierarchical taxonomic relations developed over 250 years to determine rules and conventions to catalogue species have been recently transformed into computer-based terminological resources such as NCBI taxonomy, the Thompson scientific name list, the Catalogue of Life, the Global Names Index database and the ITIS Catalogue.
Additionally, there are already available tools for the identification of species from English biomedical literature. For example, LINNAEUS (Gerner et al., 2010) and the SPECIES tool (Pafilis et al., 2013) are capable of detecting species mentions of different types. However, due to their use of rule-based systems and pattern matching, their adaptation to languages other than English is challenging. This is aggravated by the lack of resources, common evaluation scenarios, and shared tasks in other languages.
However, these resources are not fully aligned with the development of automatic systems for semantic analysis of species mentions in documents. Common challenges encountered are: name changes (obsolete species names); homonymy with commonly used words (e.g.: “Spot” refers to the species Leiostomus xanthurus or “Permit” to Trachinotus falcatus); abbreviations (sometime highly ambiguous like EC, which can be used for both for the bacterium Escherichia coli” as well as “endothelial cells” or “endometrial cancer”); incorrect cases or misspelled names (Bacterium coli, Bacillus coli and Escherichia coli for Escherichia coli); coordinations and nested expressions (“human immunodeficiency viruses types 1 and 2”); vernacular forms (common names); and role names (e.g. athletes, responders).
The LivingNER task will address these issues through a challenge on NER of species mentions and entity linking providing an exhaustively annotated large corpus of Spanish clinical case reports.
LivingNER is the first track on exhaustive species mention recognition and grounding of non-English content with a clear potential for multilingual adaptation, in particular for scientific species mentions with the aim of generating high-quality living being mention recognition components.
2. Task Summary:
LivingNER proposes the species mention recognition and normalization in Spanish clinical case reports.
Additionally, LivingNER also includes a real-world application shared task. It is a document classification task that requires providing the document class and the supporting evidence. The goal is to show that shared task-developed systems are ready to be used to solve clinical questions.
• Task 1: LivingNER-Species NER track (Species mention entity recognition): given a collection of plain text clinical case report documents, participants must return the exact character offsets of all species mentions, both human and non-human.
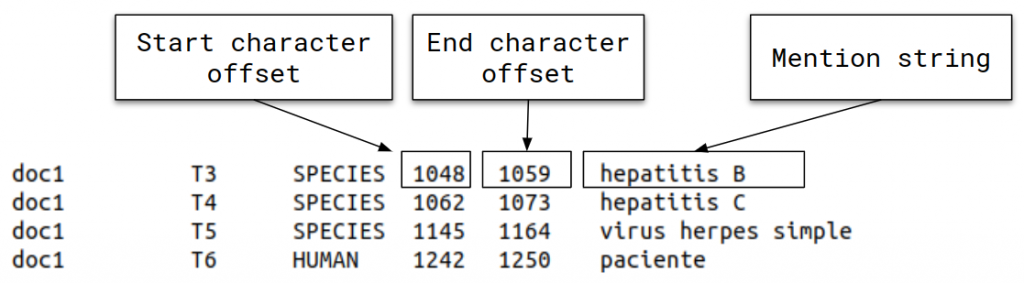
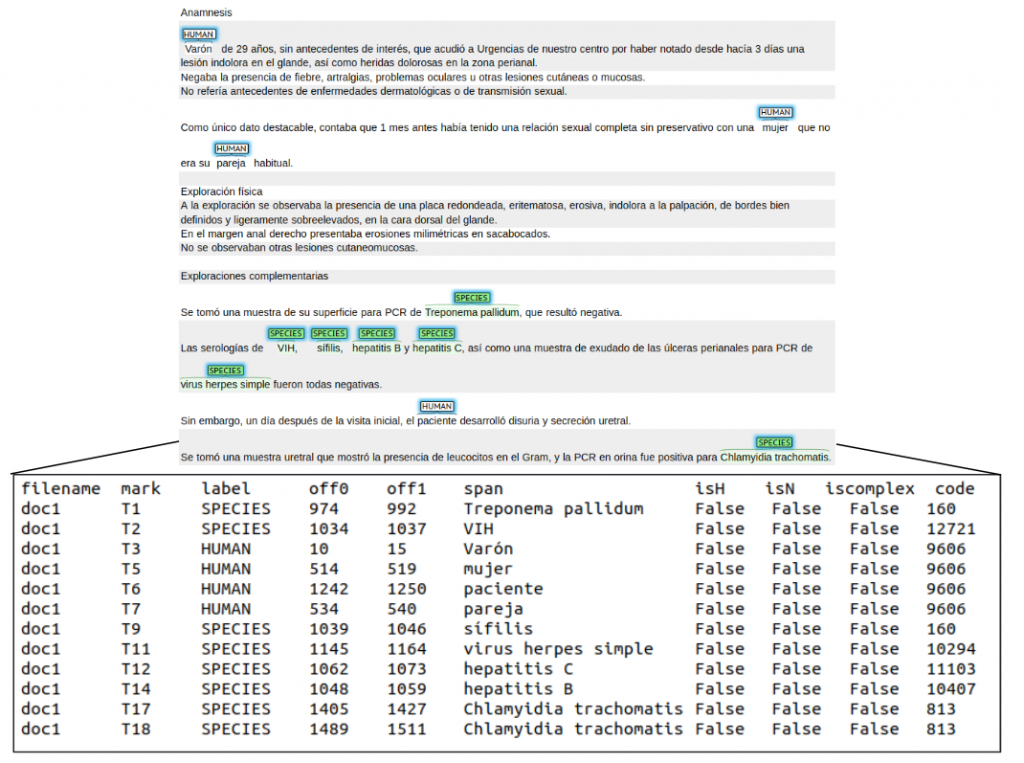
• Task 2: LivingNER-Species Norm track (Species mention normalization): given a collection of plain text clinical case report documents, participating systems have to return all species mentions together with their corresponding NCBI taxonomy concept identifiers. Find below an example of the provided dataset for this subtask:
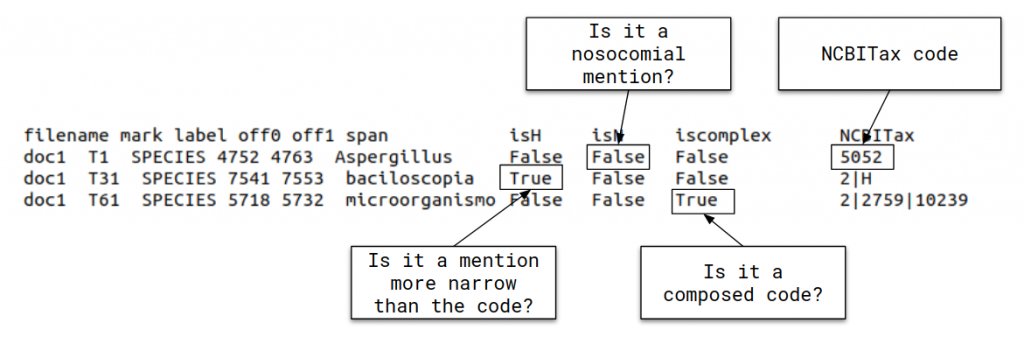
• Task 3: LivingNER-Clinical IMPACT track: given a collection of plain text documents, systems will have to:
- Perform a binary classification according to information relevant to real-world clinical use cases of high impact.
- Retrieve the list of NCBI Tax IDs that support the binary classification.
See here an example annotation for LivingNER-Clinical IMPACT track annotations:
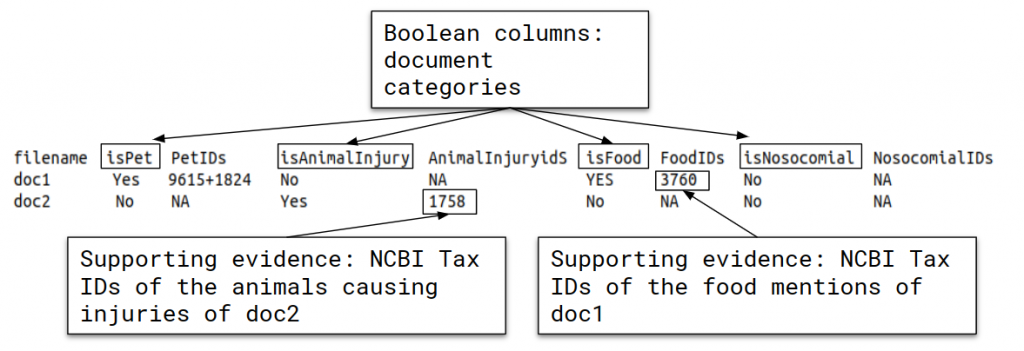
Systems have to categorize the documents in the following information axes:
- Pets and farm animals: detect whether the document contains information corresponding to pets and farm animals that live together with the patient (important for detection of animal-transmitted diseases such as toxoplasmosis, salmonellosis, cat-scratch disease, etc.). And retrieve the list of NCBI Tax IDs of the mentioned pets and farm animals.

- Animal causing injuries: detect species mentions corresponding to animals causing a direct injury on humans like bites, stings, etc. (e.g. spiders, wasps, bees). Parasites are NOT included. And retrieve the list of NCBI Tax IDs of the mentioned animal causing injuries.

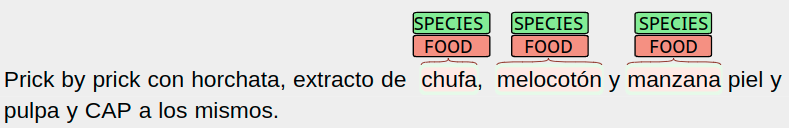
- Food species: detect all species mentions corresponding to food. This includes ingested aliments, but also any other food mentions appearing in the document. This does not include other entities intaken by the patient but are not food. And retrieve the list of NCBI Tax IDs of the mentioned food species.
- Nosocomial entities: detect species mentions corresponding to nosocomial, hospital-acquired, or healthcare-associated infections. And retrieve the list of NCBI Tax IDs of the mentioned nosocomial pathogens.

3. LivingNER Gold Standard corpus
Participation at the LivingNER track allows access to a manually annotated Gold Standard corpus generated by experts in microbiology. The quality of the dataset was assured through elaborate annotation guidelines and consistency analysis (inter-annotation agreement). The clinical cases used for the LivingNER corpus were manually selected and carefully pre-processed, in order to resemble the content of clinical records covering a diversity of medical disciplines.
Download the training, validation, test and background sets from Zenodo.
4. Scientific Publications
Participants will have the opportunity to publish their system descriptions at the IberLEF proceedings. Have a look at 2021 Meddoprof proceedings (here) and 2020 Cantemist proceedings (here).