DISTEMIST: DISease TExt Mining Shared Task
The DISTEMIST Track is promoted by Plan de Impulso de las Tecnologías del Lenguaje (Plan TL)
Generated resources
- Data: Gold Standard*, Multilingual Corpus, Silver Standard & Annotation Guidelines
- Conference Proceedings
- YouTube presentation
- Participant codes
* NEW February 7th 2023: The annotated Test Set has been publicly released
Please, cite us:
Miranda-Escalada, A., Gascó, L., Lima-López, S., Farré-Maduell, E., Estrada, D., Nentidis, A., Krithara, A., Katsimpras, G., Paliouras, G., & Krallinger, M. (2022). Overview of DisTEMIST at BioASQ: Automatic detection and normalization of diseases from clinical texts: results, methods, evaluation and multilingual resources. Working Notes of Conference and Labs of the Evaluation (CLEF) Forum. CEUR Workshop Proceedings
@article{miranda2022overview,
title={Overview of DisTEMIST at BioASQ: Automatic detection and normalization of diseases from clinical texts: results, methods, evaluation and multilingual resources},
author={Miranda-Escalada, Antonio and Gascó, Luis and Lima-López, Salvador and Farré-Maduell, Eulàlia and Estrada, Darryl and Nentidis, Anastasios and Krithara, Anastasia and Katsimpras, Georgios and Paliouras, Georgios and Krallinger, Martin},
booktitle={Working Notes of Conference and Labs of the Evaluation (CLEF) Forum. CEUR Workshop Proceedings},
year={2022}
}
Background and motivation
Semantic indexing is a good tool to improve Information Retrieval [1]. The development of biomedical semantic indexing systems has motivated several shared-tasks in the past, especially in the framework of the BioASQ workshop, achieving an outstanding impact in the field of literature in English and Spanish. These tasks have driven the improvement of automatic indexing solutions, although their evolution is gradually being limited for several reasons, including the large size of the controlled terminologies used for document indexing and the heterogeneity of the concepts, which makes it difficult to assign correct identifiers to the texts. In addition, there are some inconsistencies in manual indexing, which hinders the development of artificial intelligence systems; and the volume of training data is not large enough, especially in non-English indexing.

When running information retrieval queries, certain types of entities are particularly relevant to the search process. Focusing efforts on improving the indexing of relevant entities in the search can have a major impact on information retrieval, and can also improve the interoperability of solutions to other domains such as the clinical domain.
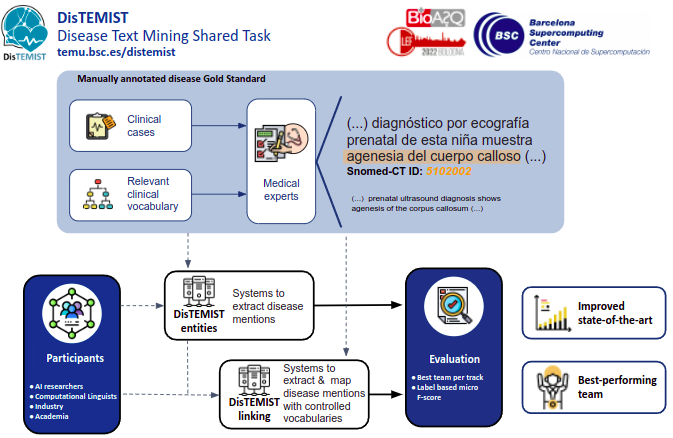
More than 20% of PubMed search queries are related to diseases, disorders, and anomalies [2]. This category has a significant presence in both scientific articles and clinical narratives[3], so the development of highly efficient systems capable of making these types of entities accessible by search systems has a great interest in the biomedical field. More granular text annotation can improve indexing systems, given that when indexing a text document, not the entire document is relevant for this purpose. In an effort to improve indexing systems by means of correct indexing relevant semantic types, the DISTEMIST track invites researchers, biomedical industry professionals, NLP, and ontology experts to develop systems capable of indexing the content about diseases in biomedical texts, basing the choice on the existing evidence in the text by using NER, entity linking, and cross-ontology mapping techniques.
Task Summary
The DISTEMIST will be structured into two independent sub-tasks, each taking into account a particularly important use case scenario:
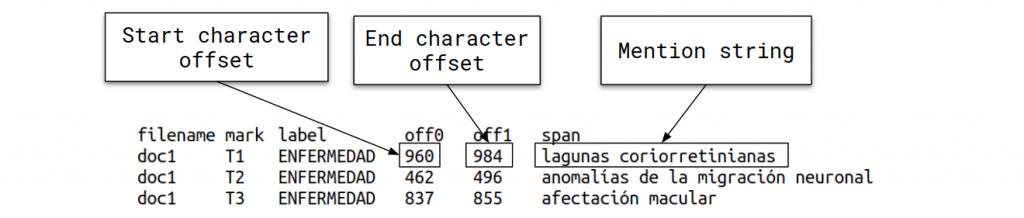
- DISTEMIST-entities subtrack: requires automatically finding disease mentions in published clinical cases.
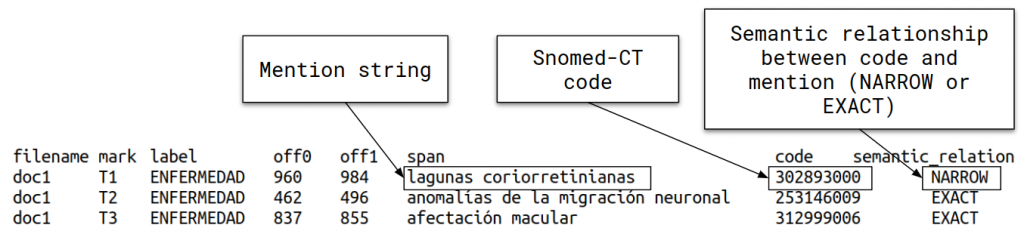
- DISTEMIST-linking subtrack: requires automatically finding disease mentions in published clinical cases and assigning, to each mention, a Snomed-CT term.
For this task, we have prepared a set of corpora that can be examined deeply in the data section.
References
[1]. Nentidis, A., Katsimpras, G., Vandorou, E., Krithara, A., Gasco, L., Krallinger, M., & Paliouras, G. (2021, September). Overview of BioASQ 2021: The Ninth BioASQ Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering. In International Conference of the Cross-Language Evaluation Forum for European Languages (pp. 239-263). Springer, Cham.
[2]. Islamaj Dogan, R., Murray, G. C., Névéol, A., & Lu, Z. (2009). Understanding PubMed® user search behavior through log analysis. Database, 2009.
[3]. Gasco, L., Nentidis, A., Krithara, A., Estrada-Zavala, D., Murasaki, R. T., Primo-Peña, E., … & Krallinger, M. (2021). Overview of BioASQ 2021-MESINESP track. Evaluation of advance hierarchical classification techniques for scientific literature, patents and clinical trials. CEUR Workshop Proceedings.
Schedule
| Event | Date (Midnight CEST) |
|---|---|
| Sub-track 1: DISTEMIST-entities Train Set Release | April, 1 |
| Sub-track 2: DISTEMIST-linking Train Set Release 1 | April, 13 |
| Release of DISTEMIST annotation guidelines | April, 22 |
| Sub-track 2: DISTEMIST-linking Train Set Release 2 | April, 25 |
| Test Set Release (DISTEMIST-entities and linking) | May, 10 |
| Participant Test Prediction Due (DISTEMIST-entities and linking) | May, 21 ("Anywhere on Earth") |
| Participant evaluation result release | May, 25 |
| Working Notes deadline | June, 2 UPDATED |
| Deadline for reviews & Notification of acceptance of participant papers | June, 20 UPDATED |
| Deadline for submission of camera-ready participant papers | July, 1 |
| BioASQ @ CLEF2022 | September, 5-8 September |